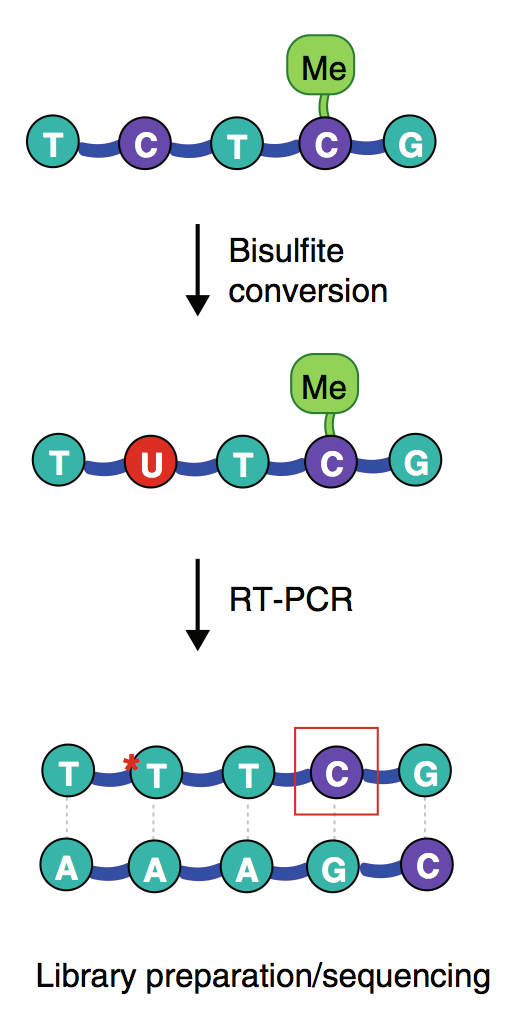
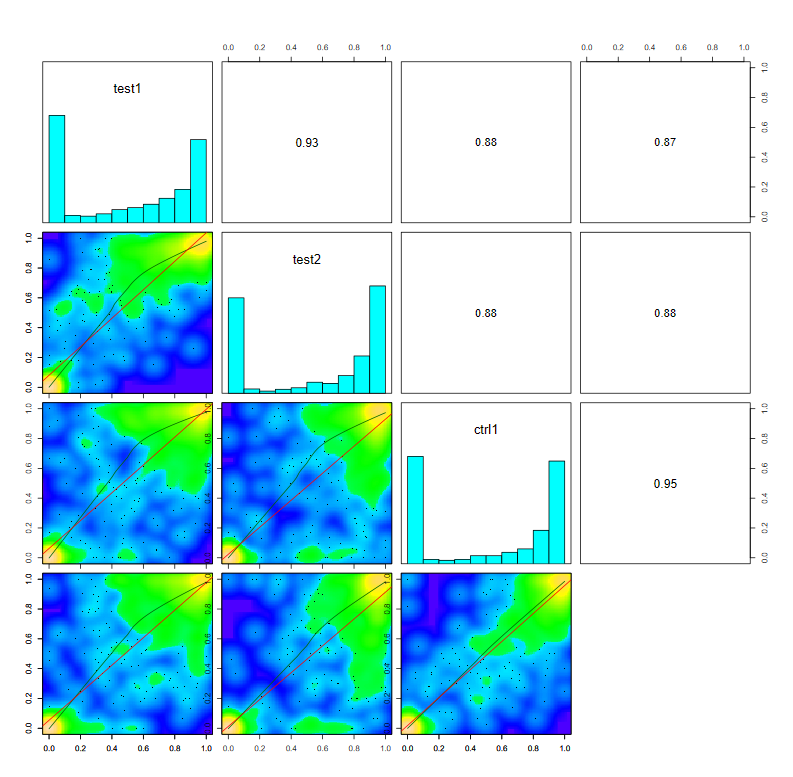
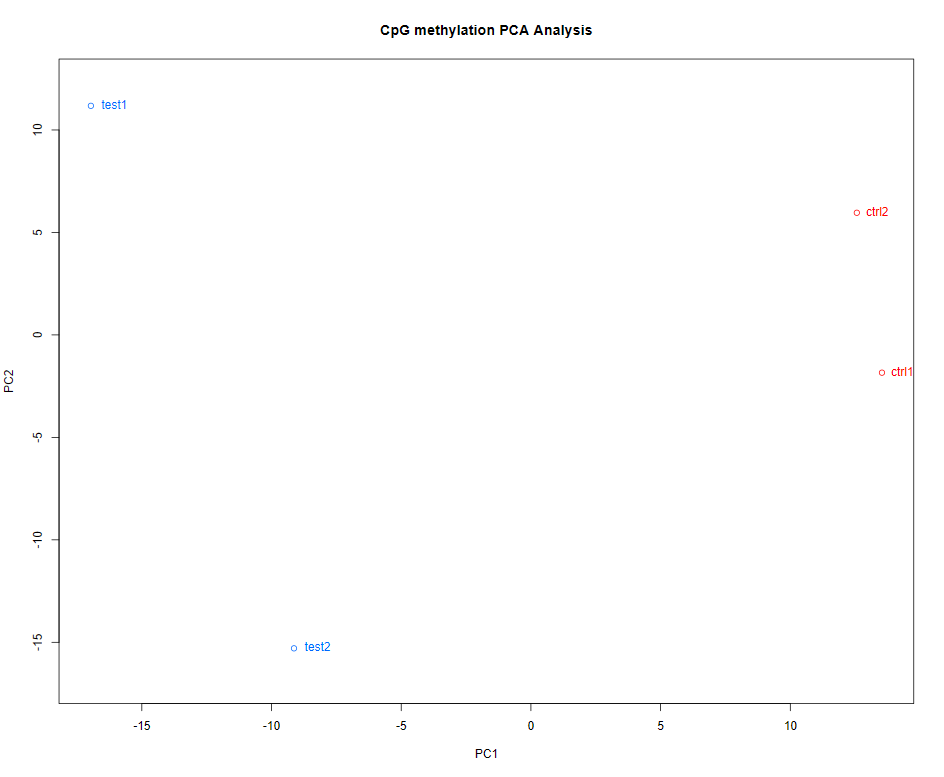
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
|
files_list <- list(
"13.sam4methkit/WT1.sorted.sam", "13.sam4methkit/WT2.sorted.sam", "13.sam4methkit/WT3.sorted.sam",
"13.sam4methkit/YS1.sorted.sam", "13.sam4methkit/YS2.sorted.sam", "13.sam4methkit/YS3.sorted.sam",
"13.sam4methkit/RS1.sorted.sam", "13.sam4methkit/RS2.sorted.sam", "13.sam4methkit/RS3.sorted.sam"
)
methylKit::processBismarkAln(
location = files_list,
sample.id = list("WT1", "WT2", "WT3", "YS1", "YS2", "YS3", "RS1", "RS2", "RS3"),
assembly = "C88",
save.folder = "14.methykit",
read.context = "CpG",
save.context = "CpG",
treatment = c(0, 0, 0, 1, 1, 1, 2, 2, 2)
) -> obj.cpg
save(obj.cpg, file = "14.methykit/CpG.RData")
|